FME
Food Microbial Ecology
Understanding the microbial ecology of the food chain: towards more sustainable and healthy food.
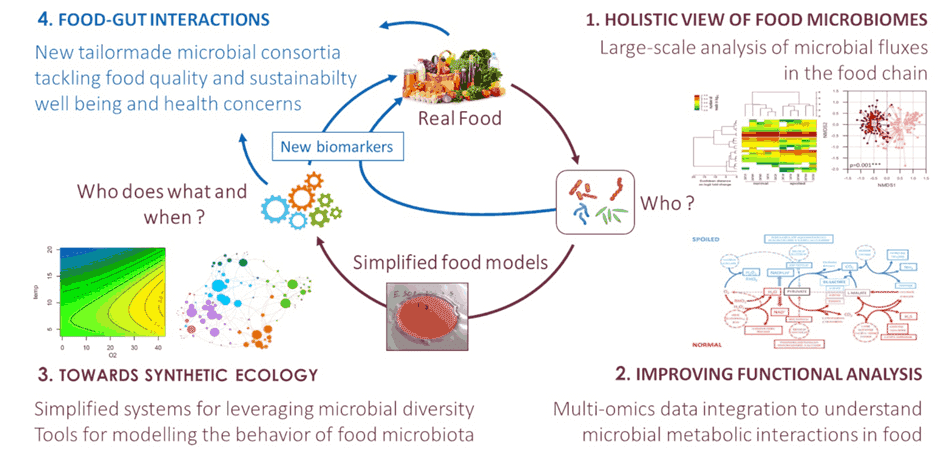
The FME lab focuses its research on understanding the mechanisms by which bacteria and fungi interact with food products during spoilage and fermentation. In particular, our team is trying to understand how food microbial diversity may be leveraged to design microbial solutions for more sustainable and healthy fermented food production. We develop fundamental analysis of food microbial ecology, including metagenomics, metatranscriptomics and metabolomics, comparative genomics and global analyses of the metabolic functions carried out by the different microbial species. As shown in the figure, this scientific strategy is based on four research axes. A strongly inter-disciplinary strategy combines results and approaches from microbial ecology, high throughput omics data analysis, and computational biology. Our scientific activity creates a strong foreground to develop downstream innovative programs with industrial stakeholders in fermented food areas. The FME lab is also managing the Ferment-du-Futur BioLector XT micro-fermenters in the frame of the PIAM platform (link to the page) for studying metabolic microbial interactions within food models. More news about our lab and our various activities?

Marie Champomier-Vergès & Stéphane Chaillou
Research axis
Holistic view of food microbiomes in the food chain
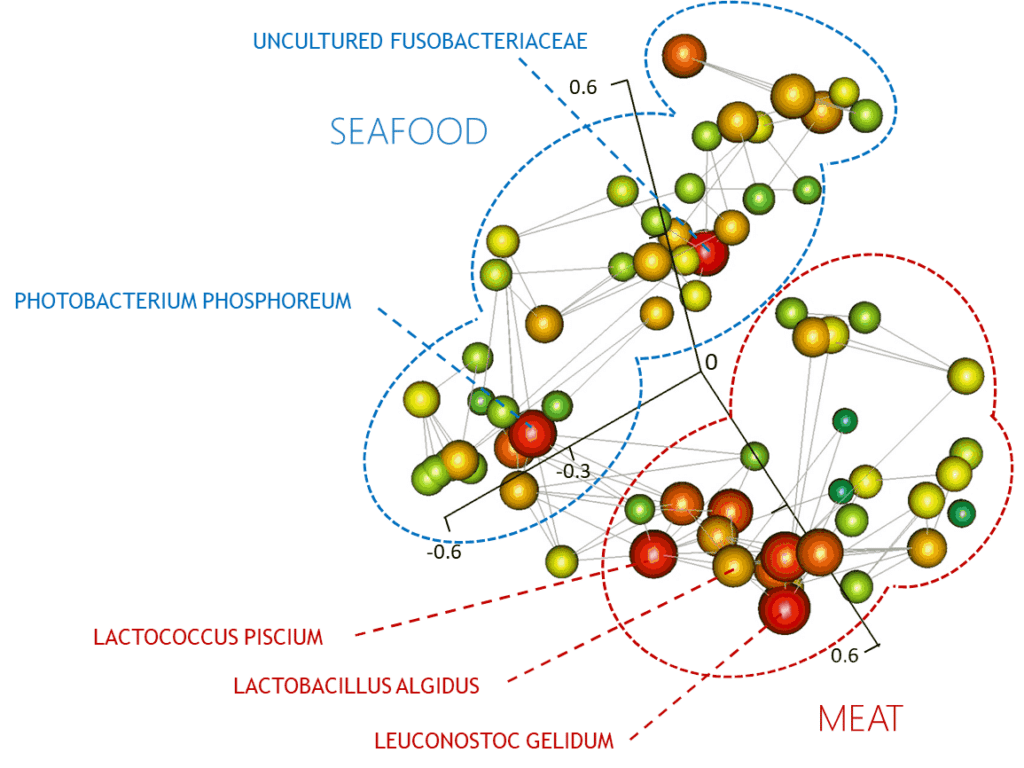
Our research uses two approaches. The first of these consists of large and ambitious sampling surveys to collect from hundreds to thousands of data points on given industrial environment or type of food. Metagenetic analysis (for example with 16S rRNA genes, ITS or gyrB markers) and multivariate discriminant analysis are then performed to unravel how the food production parameters shape the microbial communities. Our work has demonstrated the potential of this type of data (obtained in collaboration with industrial partners) for the development of predictive tools and in characterization of the processing steps that can influence the microbial composition and activity in the context of food production and storage. The second approach has as its basis a thorough functional analysis of food microbiomes combined using shotgun metagenomics and comparative genomic and functional analysis of new strains and species isolated from food. In this context, we are setting up an easy-access metagenomic study tool integrating public genomic data (genes, functions …) with metagenomic data (gene profiles and nucleotide variations inferred from shotgun sequencing) and ecological and technological datasets.
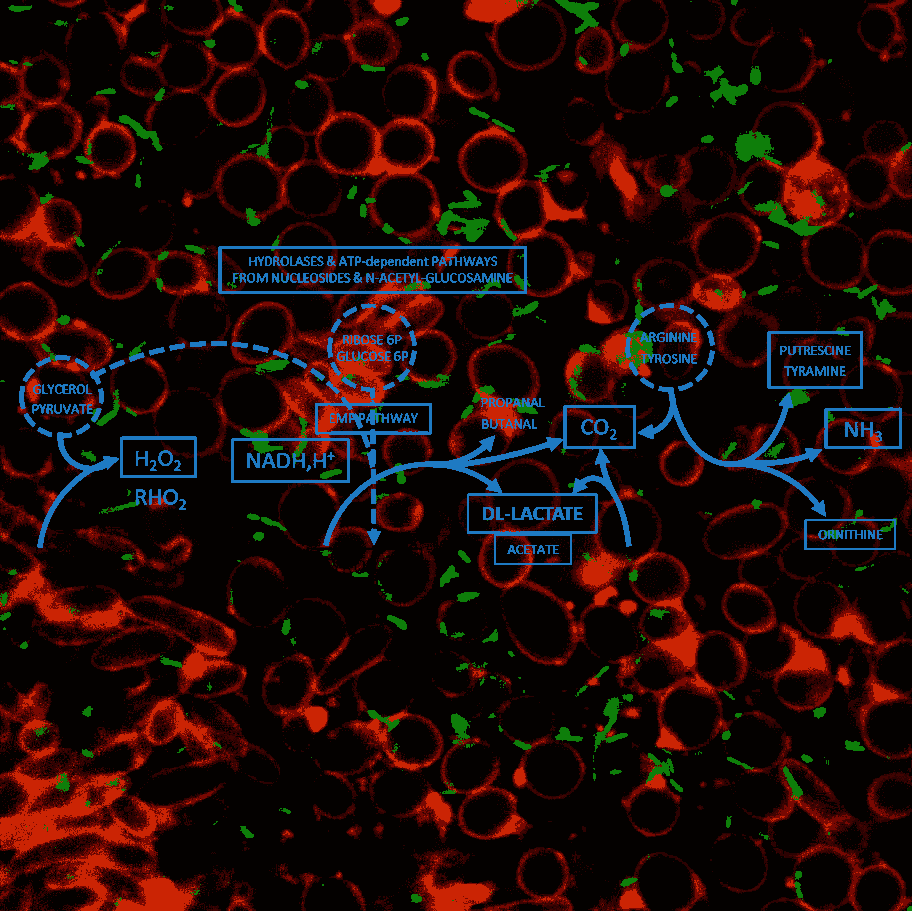
Improving functional analysis of metabolic interactions in food
Towards synthetic food microbial ecology
The object of this axis to develop new microbial consortia to address health or sustainability issues by taking advantage of knowledge gained on microbial interactions and strains genomic sequences. To this end we are developing strategies which will allow us to verify the observations made “in alimentariis” and the hypotheses built from these data on metabolic interactions. This involves the design of simplified food systems and the development of tools for modelling the behaviour of artificial microbial consortia. Our “synthetic ecology approach” aims to elaborate hypotheses on the possible metabolic interactions within a microbial community, and to build an experimental approach whose objective is to test them. It is, therefore, a repetitive, iterative strategy whose results are used to reformulate and refine the hypotheses which are then retested experimentally, the aim being always to improve the level of understanding of the interactions when compared to data obtained from real-life food ecosystems.
Food-Gut interactions

Food microbes can have major positive, or negative, impacts on the production of various fermented foods, and in the human gut and digestive health. Although a broad diversity of microbial communities can be identified across many fermented foods, significant information about their contribution to human health is lacking. We aim to understand how diversity of dietary microbes, contribute to a healthy gut microbiome and how the long-term consumption of fermented food contributes to a beneficial health impact.
Our objective is to use food and gut microbiome data to allow a more relevant health-orientated design of microbial consortia for fermented food and to identify biomarkers related to the health effect of these fermented foods.
Team members
Alumni
- Liste à puce
- Liste à puce
- Liste à puce
- Liste à puce
- Liste à puce
- Liste à puce
- Liste à puce
Key points
- Poirier, S. et al. Holistic integration of omics data reveals the drivers that shape the ecology of microbial meat spoilage scenarios. Frontiers in Microbiology 14, 1286661 (2023).
- Kothe, C. I., Bolotin, A., Kraïem, B.-F., Dridi, B. & Renault, P. Unraveling the world of halophilic and halotolerant bacteria in cheese by combining cultural, genomic and metagenomic approaches. International journal of food microbiology 358, 109312 (2021).
- Zagdoun, M., Coeuret, G., N’Dione, M., Champomier-Vergès, M.-C. & Chaillou, S. Large microbiota survey reveals how the microbial ecology of cooked ham is shaped by different processing steps. Food Microbiology 91, 103547 (2020).
- Verplaetse, E. et al. Heme Uptake in Lactobacillus sakei Evidenced by a New Energy Coupling Factor (ECF)-Like Transport System. Applied and environmental microbiology 86, (2020).
- Terán, L. C. et al. Phylogenomic Analysis of Lactobacillus curvatus Reveals Two Lineages Distinguished by Genes for Fermenting Plant-Derived Carbohydrates. Genome biology and evolution 10, 1516–1525 (2018).
- Chaillou, S. et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. The ISME journal 9, 1105–1118 (2015).
All the team publications are available in the FME-MICALIS HAL collection.
- Horizon Europe RIA DOMINO (coordination). Harnessing the microbial potential of fermented food for healthy & sustainable food systems. 2023-2028.
- ANR-21-CE21-0003-01 METASIMFOOD (coordination). Improving the quality, safety and sustainability of fermented vegetable food and fruit beverage using a knowledge-driven synthetic ecology and modelling approach. 2022-2025.
- ANR-16-CE21-0006-03 REDLOSSES (partner). REDucing food LOSSES by microbial spoilage prediction. 2016-2020.
- ANR-14-CE20-0004-01 BLACHP (Partner). LACtic Bacteria combined with High Pressure for a sustainable stabilization process of refrigerated meat products. 2015-2018.
- ANR-ALIA ECOBIOPRO (coordination). Exploration of microbial ecosystems of fish and meat products. Effects of bioprotective cultures. 2010-2014.
- FERMENT DU FUTUR AAP2023 SYNTHPLEX 2023-2025 (coordination).
- FERMENT DU FUTUR AAP2023 SYNERGY 2023-2025 (coordination).
- HOLOFLUX metaprogramme EGG-TO-MEAT 2020-2021 (partner)
- DIGIT-BIO metaprogramme FERMENTWIN 2024-2025 (partner)
- MEM metaprogramme VIROME ACCESS 2018-2020 (coordination)
- MEM metaprogramme COMETES 2010-2012 (coordination)
- EMOVOL (private) 2021-2025 (coordination).