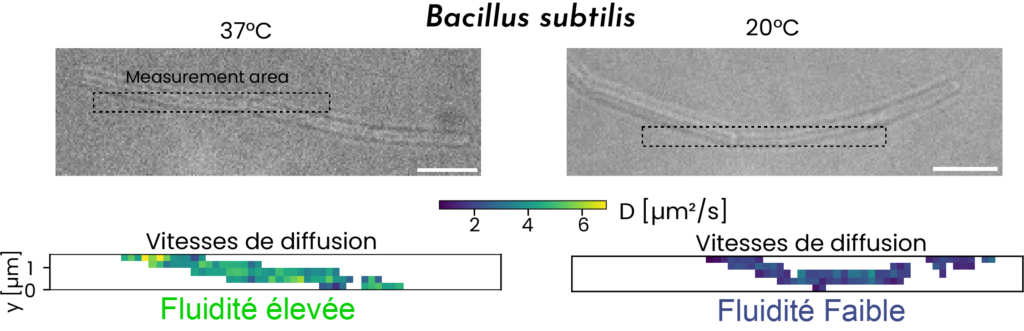
- Barbotin A, Billaudeau C, Sezgin E, Carballido-López R. Quantification of membrane fluidity in bacteria using TIR-FCS. Biophys J. 2024. doi: 10.1101/2023.10.13.562271
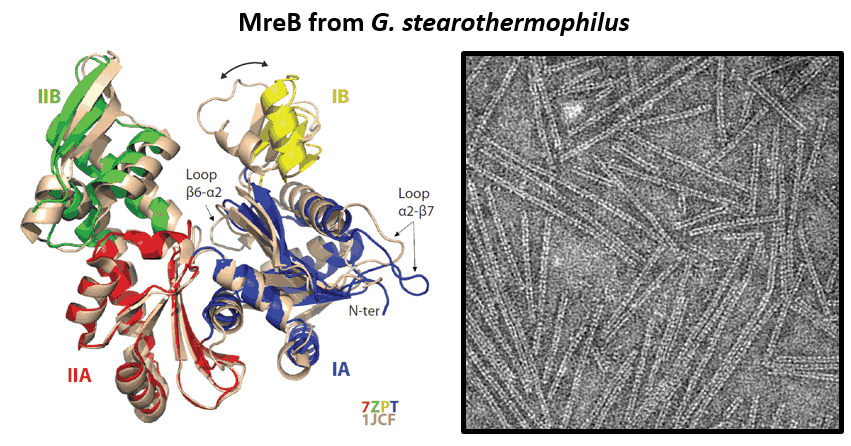
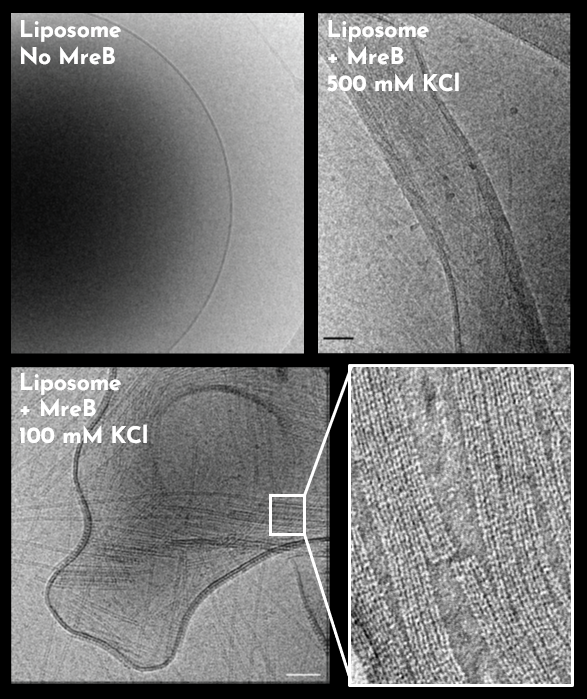
- Mao W, Renner L, Cornilleau C, Li de la Sierra-Gallay I, Afensiss S, Benlamara S, Ah-Seng Y, Van Tilbeurgh H, Nessler S, Bertin A, Chastanet A, Carballido-Lopez R. On the role of nucleotides and lipids in the polymerization of the actin homolog MreB from a Gram-positive bacterium. elife. 2023 Oct 11:12:e84505. doi: 10.7554/eLife.84505.
- Labarde A, Jakutyte L, Billaudeau C, Fauler B, López-Sanz M, Ponien P, Jacquet E, Mielke T, Ayora S, Carballido-López R, Tavares P. Temporal compartmentalization of viral infection in bacterial cells. Proc Natl Acad Sci U S A. 2021 Jul 13;118(28):e2018297118. doi: 10.1073/pnas.2018297118.
- Billaudeau C, Yao Z, Cornilleau C, Carballido-Lopez R, Chastanet A. MreB Forms Subdiffraction Nanofilaments during Active Growth in Bacillus subtilis. MBio. 2019 Jan 29; doi: 10.1128/mBio.01879-18.
- Krokowski S, Lobato-Márquez D, Chastanet A, Pereira PM, Angelis D, Galea D, Larrouy-Maumus G, Henriques R, Spiliotis ET, Carballido-López R, Mostowy S. Septins Recognize and Entrap Dividing Bacterial Cells for Delivery to Lysosomes. Cell Host Microbe. 2018 Dec 12; doi: 10.1016/j.chom.2018.11.005.
- Mirouze N, Ferret C, Cornilleau C, Carballido-López R. Antibiotic sensitivity reveals that wall teichoic acids mediate DNA binding during competence in Bacillus subtilis. Nat Commun. 2018 Nov 29; doi: 10.1038/s41467-018-07553-8.
- Billaudeau C, Chastanet A, Yao Z, Cornilleau C, Mirouze N, Fromion V, Carballido-Lopez R. Contrasting mechanisms of growth in two model rod-shaped bacteria. Nat Commun. 2017 Jun 7; doi: 10.1038/ncomms15370.
- Mirouze N, Ferret C, Yao Z, Chastanet A, Carballido-López R. MreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in Bacillus subtilis. PLoS Genet. 2015 Jun 19; doi: 10.1371/journal.pgen.1005299
- Yao Z, Carballido-López R. Fluorescence imaging for bacterial cell biology: from localization to dynamics, from ensembles to single molecules. Annu Rev Microbiol. 2014;68:459-76. doi: 10.1146/annurev-micro-091213-113034
- Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Söldner R, Carballido-López R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011 Jul 8; doi: 10.1126/science.1203466
For a full list see here.