- Direct comparison of spatial transcriptional heterogeneity across diverse Bacillus subtilis biofilm communities. Dergham Y, Le Coq D, Nicolas P, Bidnenko E, Dérozier S, Deforet M, Huillet E, Sanchez-Vizuete P, Deschamps J, Hamze K, Briandet R. Nat Commun. 2023 Nov 20;14(1):7546. doi: 10.1038/s41467-023-43386-w. PMID: 37985771
- Combining Fusion of Cells with CRISPR-Cas9 Editing for the Cloning of Large DNA Fragments or Complete Bacterial Genomes in Yeast. Guesdon G, Gourgues G, Rideau F, Ipoutcha T, Manso-Silván L, Jules M, Sirand-Pugnet P, Blanchard A, Lartigue C. ACS Synth Biol. 2023 Nov 17;12(11):3252-3266. doi: 10.1021/acssynbio.3c00248. PMID: 37843014
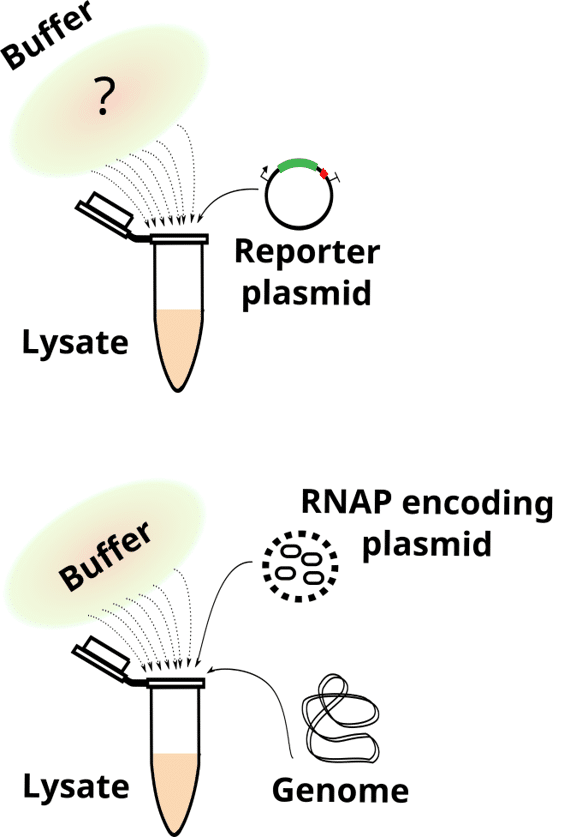
- What remains from living cells in bacterial lysate-based cell-free systems. Wagner L, Jules M, Borkowski O. Comput Struct Biotechnol J. 2023 May 24;21:3173-3182. doi: 10.1016/j.csbj.2023.05.025. eCollection 2023. PMID: 37333859.
- Greedy reduction of Bacillus subtilis genome yields emergent phenotypes of high resistance to a DNA damaging agent and low evolvability. Dervyn E, Planson AG, Tanaka K, Chubukov V, Guérin C, Derozier S, Lecointe F, Sauer U, Yoshida KI, Nicolas P, Noirot P, Jules M. Nucleic Acids Res. 2023 Mar 15:gkad145. doi: 10.1093/nar/gkad145. PMID: 36919610.
- Termination factor Rho mediates transcriptional reprogramming of Bacillus subtilis stationary phase. Bidnenko V, Nicolas P, Guérin C, Dérozier S, Chastanet A, Dairou J, Redko-Hamel Y, Jules M, Bidnenko E. PLoS Genet. 2023 Feb 3;19(2):e1010618. doi: 10.1371/journal.pgen.1010618. eCollection 2023 Feb. PMID: 36735730.
- Expanding Diversity of Firmicutes Single-Strand Annealing Proteins: A Putative Role of Bacteriophage-Host Arms Race. Steczkiewicz K, Prestel E, Bidnenko E, Szczepankowska AK. Front Microbiol. 2021 Apr 20;12:644622. doi: 10.3389/fmicb.2021.644622. eCollection 2021. PMID: 33959107
- SppI Forms a Membrane Protein Complex with SppA and Inhibits Its Protease Activity in Bacillus subtilis. Henriques G, McGovern S, Neef J, Antelo-Varela M, Götz F, Otto A, Becher D, van Dijl JM, Jules M, Delumeau O. mSphere. 2020 Oct 7;5(5):e00724-20. doi: 10.1128/mSphere.00724-20. PMID: 33028682.
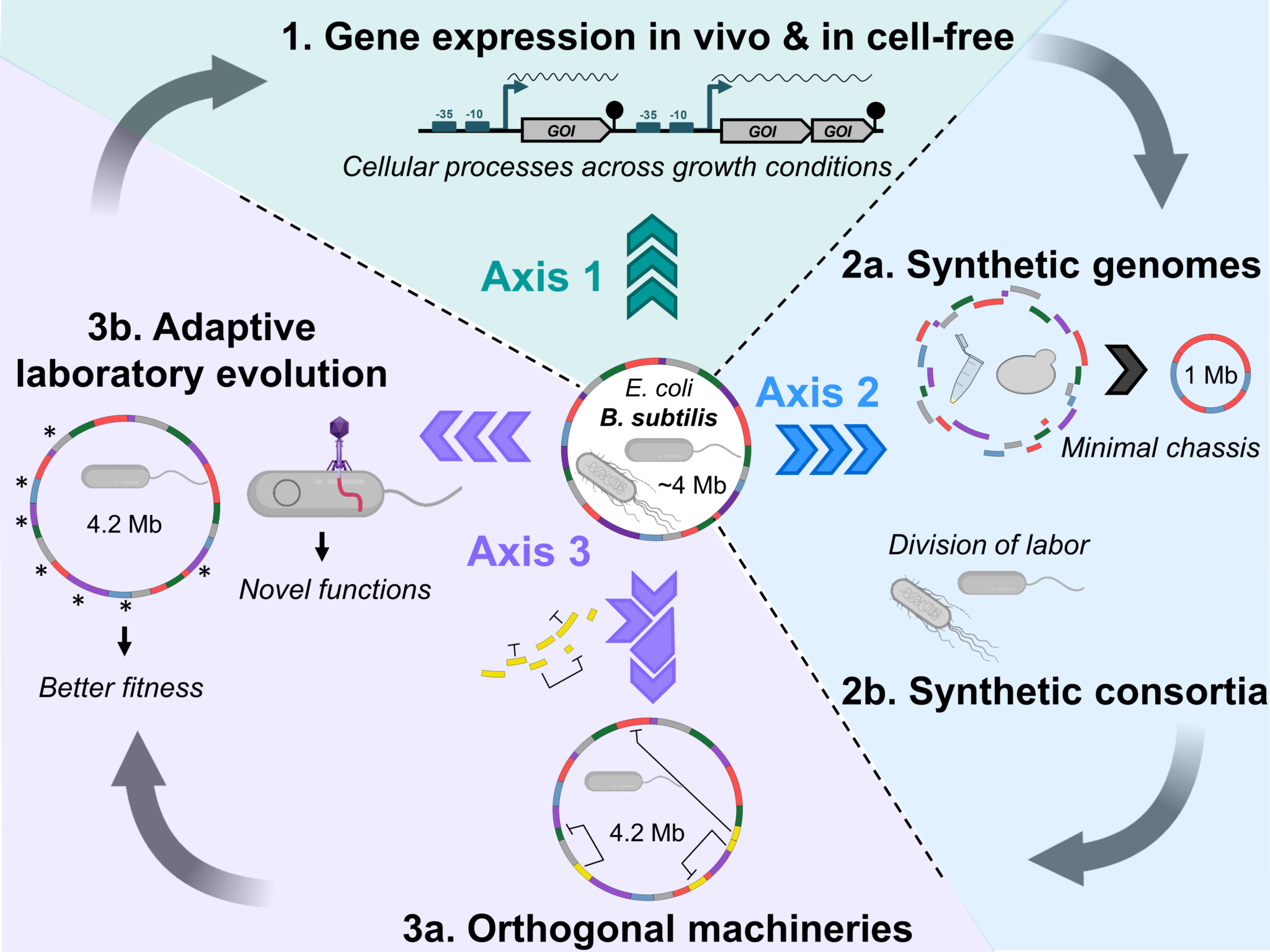
- Bacterial growth physiology and RNA metabolism. Planson AG, Sauveplane V, Dervyn E, Jules M. Biochim Biophys Acta Gene Regul Mech. 2020 May;1863(5):194502. doi: 10.1016/j.bbagrm.2020.194502. PMID: 32044462 Review.
All the team publications are available in the SYBER-MICALIS HAL collection.