The INRAE Micalis Institute and Lallemand Animal Nutrition (LAN) are pleased to announce the creation of the LabCom “Biofilm1Health,” a collaborative laboratory focused on unlocking the potential of biofilm properties in beneficial microbes. This cutting-edge partnership between the B3D team from the Micalis Institute (UMR 1319 INRAE, AgroParisTech, Université Paris-Saclay) and LAN, with support from the French National Research Agency (ANR), aims to advance the One Health concept, a holistic approach that integrates human, animal, and environmental health.
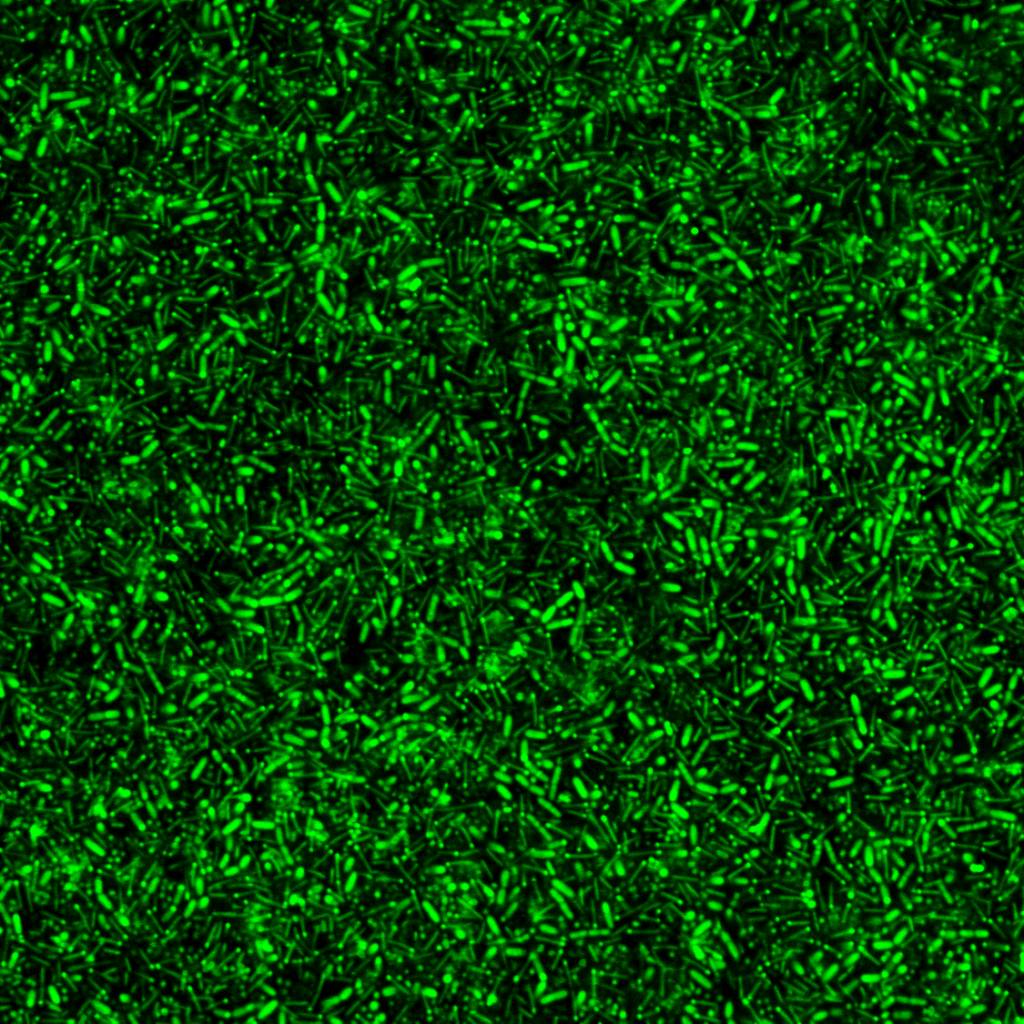
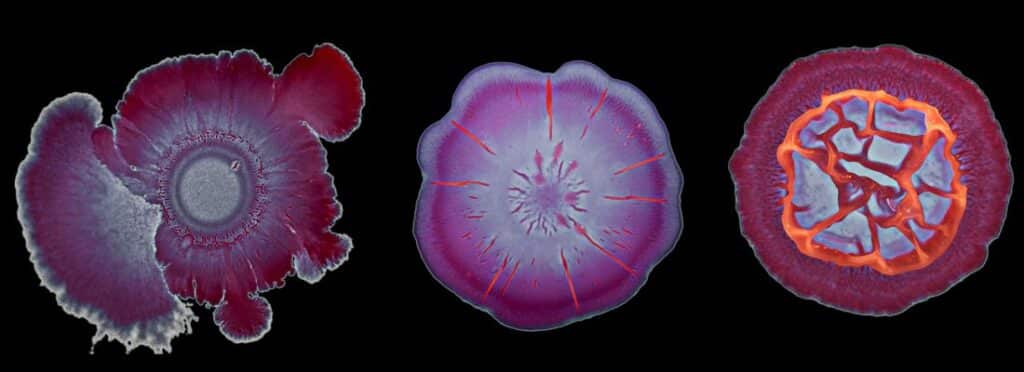
The mission of LabCom “Biofilm1Health” is to explore the unique properties of beneficial biofilms—complex microbial communities that offer protection and enhanced functionality. By investigating how these biofilms can reduce harmful microorganisms in livestock, food production, and the environment, the initiative seeks to provide sustainable and effective microbial solutions. A defining aspect of this project is its incorporation of advanced biofilm phenotyping and artificial intelligence to enhance our understanding of these intricate ecosystems.
The ultimate goal is to optimize the application of beneficial biofilms to create next-generation solutions that improve sustainability, reduce the need for chemical interventions, and support animal welfare.
The project is structured around four key research axes:
- Axis 1 : Selection of beneficial microbial strains and consortia that antagonize undesirable microorganisms in biofilms
- Axis 2 : Detail investigation into microbial interaction mechanisms within multi-species biofilms
- Axis 3 : Optimization of product formulation for enhanced field applications
- Axis 4 : Development of in-situ methodologies for studying environmental biofilms
A strategic partnership driving scientific and industrial innovation
The B3D team at the Micalis Institute is internationally recognized for its pioneering research on biofilms and spatially organized microbial communities. Their work focuses on understanding the complex relationships between microbial community structures and their emergent properties, particularly in biofilm and food-related environments. By exploring phenotypic heterogeneity and interspecies interactions, the B3D team aims to develop novel preventive and curative strategies that can be applied to industries ranging from food production to healthcare and aerospace.
Lallemand Animal Nutrition has been at the forefront of microbial solutions for animal nutrition since the 1980s, including the introduction of the first registered probiotics in Europe for ruminant and monogastric animal feeds. Today, LAN plays a leading role in animal nutrition and environmental management in sectors such as ruminant, pig, poultry, aquaculture, equine, and pet nutrition. Through strategic collaborations and its Blagnac laboratory, LAN continues to innovate by developing products that harness beneficial microorganisms for both nutrition and livestock management.
Looking Ahead: A new era of microbial innovation
The creation of LabCom “Biofilm1Health” signals the dawn of a new chapter in microbial innovation, where the combined expertise of INRAE and Lallemand Animal Nutrition will drive forward solutions that benefit human, animal, and environmental health. By unlocking the potential of biofilms and leveraging cutting-edge digital tools, this collaboration will contribute to sustainable practices across multiple industries. The research carried out at LabCom “Biofilm1Health” promises to not only deepen our understanding of microbial ecosystems but also pave the way for transformative solutions that reduce chemical inputs, enhance animal welfare, and promote a healthier planet.
LabCom “Biofilm1Health” is funded by the French National Research Agency .