- Rul F, Béra-Maillet C, Champomier-Vergès MC, El-Mecherfi KE, Foligné B, Michalski MC, Milenkovic D, Savary-Auzeloux I. (2022). Underlying evidence for the health benefits of fermented foods in humans. Food Funct 10;13(9):4804-4824. doi: 10.1039/d1fo03989j.
- Boulay M, Al Haddad M, Rul F. (2020). Streptococcus thermophilus growth in soya milk: Sucrose consumption, nitrogen metabolism, soya protein hydrolysis and role of the cell-wall protease PrtS. Int J Food Microbiol. Dec 16;335:108903. doi: 10.1016/j.ijfoodmicro.2020.108903.
- Proust L, Haudebourg E, Sourabié A, Pedersen M, Besançon I, Monnet V, Juillard V (2020) Multi-omics approach reveals how yeast extract peptides shape Streptococcus thermophilus metabolism. Appl Environ Microbiol 86, e01446-20. doi: 10.1128/ AEM.01446-20.
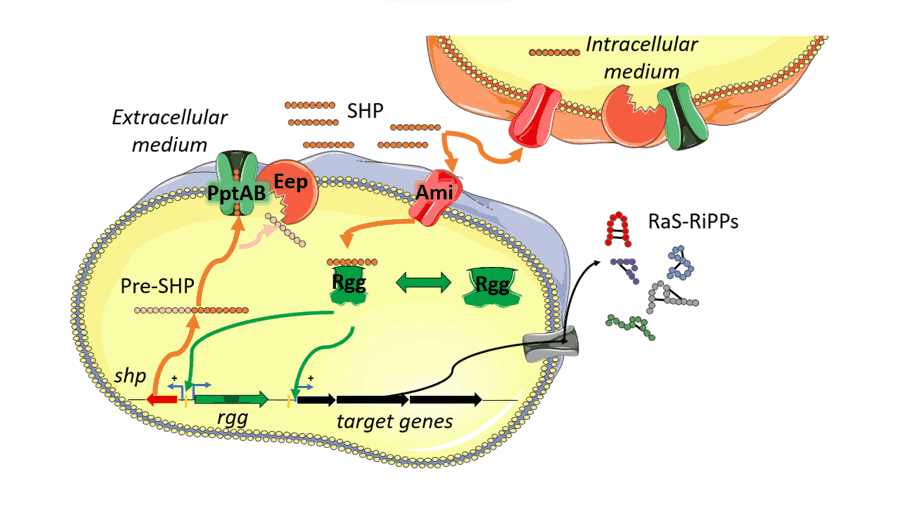
- Lingeswaran A, Metton C, Henry C, Monnet V, Juillard V, Gardan R (2020) Export of Rgg quorum sensing peptides is mediated by the pptAB ABC transporter in Streptococcus thermophilus strain LMD-9. Genes 11, 1096. doi: 10.3390/genes11091096.
- Guillot A, Boulay M, Chambellon E, Gitton C, Monnet V, Juillard V (2016) Mass spectrometry analysis of the extracellular peptidome of Lactococcus lactis: lines of evidence for the co-existence of extracellular protein hydrolysis and intracellular peptide excretion. J Prot Res 15, 3214-3224. doi: 10.1021/acs.jproteome.6b00424
- Couvigny B., Kulakauskas S., Pons N., Quinquis B., Abraham A.L., Meylheuc T., Delorme C., Renault P., Briandet R., Lapaque N., Guedon E. (2018). Identification of new factors modulating adhesion abilities of the pioneer commensal bacterium Streptococcus salivarius. Front. Microbiol. 9, 273. doi: 10.3389/fmicb.2018.00273.
All the team publications are available in the COMBAC-MICALIS HAL collection