FInE
Functionality of the Intestinal Ecosystem
The human intestinal microbiota, as we know it today, appears to be very complex. In order to move towards a holistic understanding of the gut microbiota, including its non-cultivable component, which is still in the majority to date, we have contributed to the development of a new and powerful approach, metagenomics, which has enabled rapid progress in the characterisation of the genomic and genetic diversity of the gut microbiota. For example, the microbial genome (microbiome) of each individual contains ~28 times more genes than the human genome. The microbiota is, moreover, considered to be a host organ in its own right.
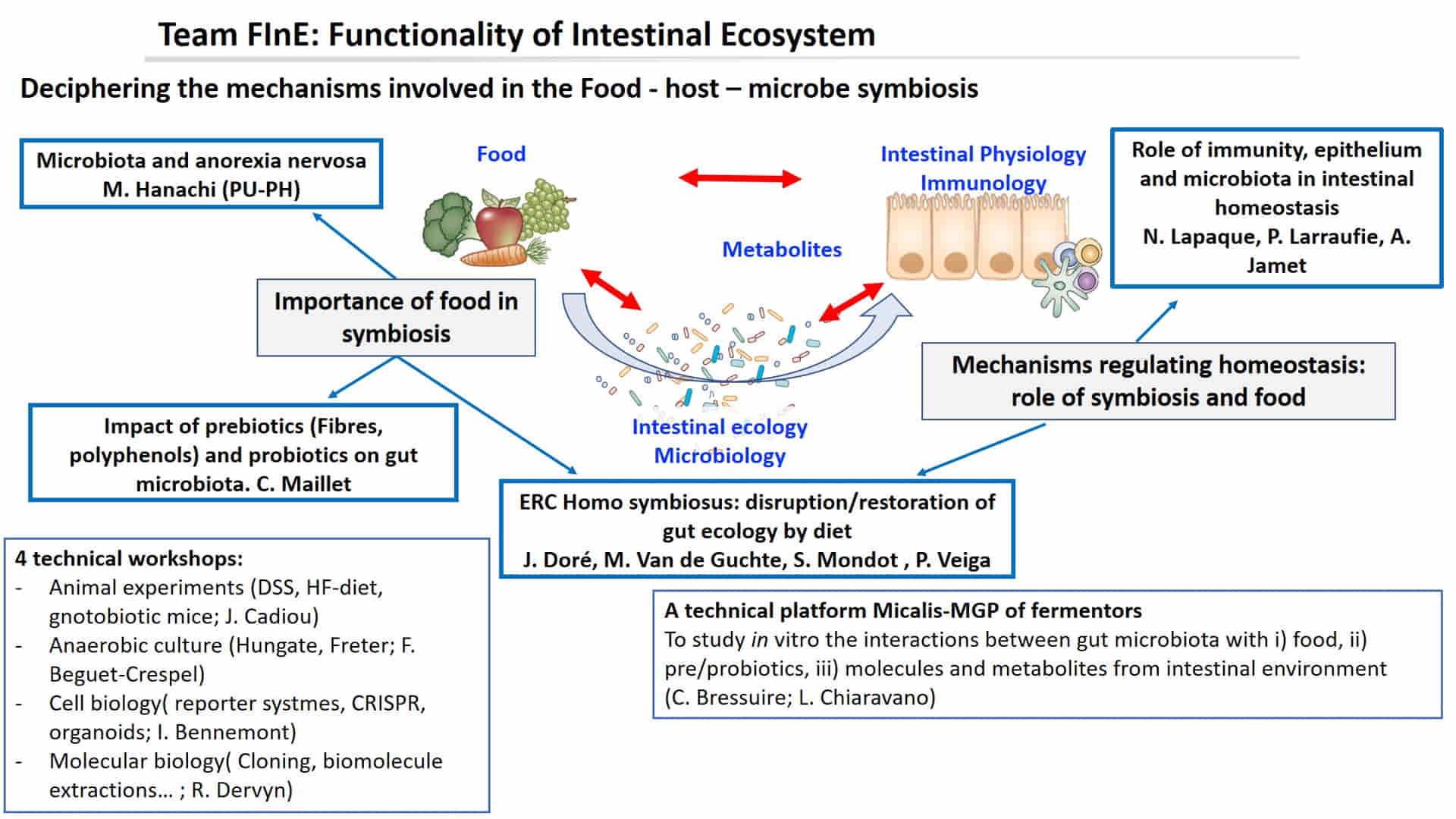
The FInE team aims to deepen the current understanding of the fundamental role played by the microbiota in human health. Our objective is to better understand the mechanisms of the human-microbe symbiosis and the influence of our diet, in particular the importance of fibre, polyphenols and probiotics, in order to define strategies for its modulation. FInE is a joint team linked to the MICA and AlimH departments of INRAE. It is closely linked to MetaGenoPolis since it contributed to the emergence of two of its platforms (SAMBO and MétaFun).
Research axis
Anorexia nervosa and microbiota
Impact of food, probiotics… on intestinal ecology
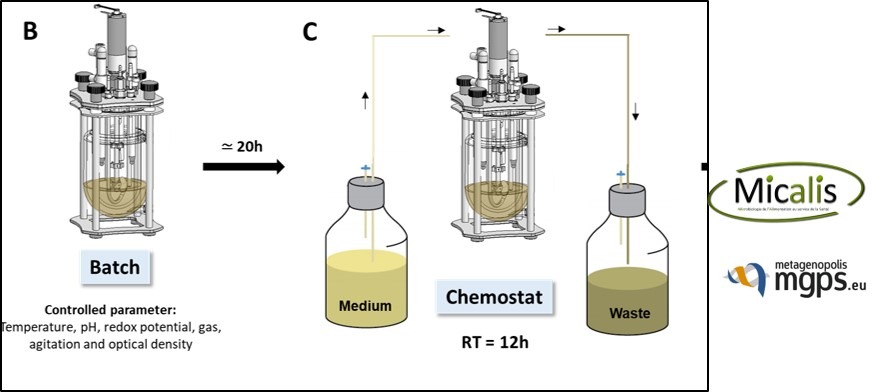
- Diversity richness (16S rRNA), metabolites (SCFA…), quantitative metagenomics, test on epithelial cell reporter system and enzymatic assays, evolution of bacteria (pathogen or commensal)
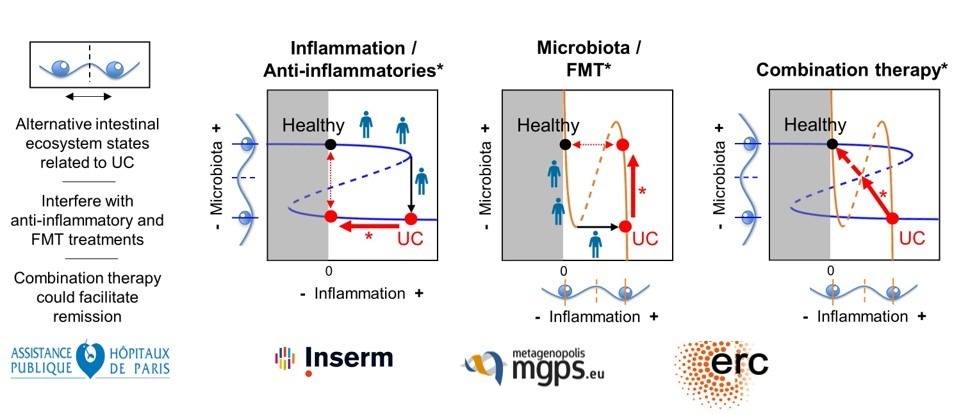
A new concept of critical transition in the intestinal ecosystem.
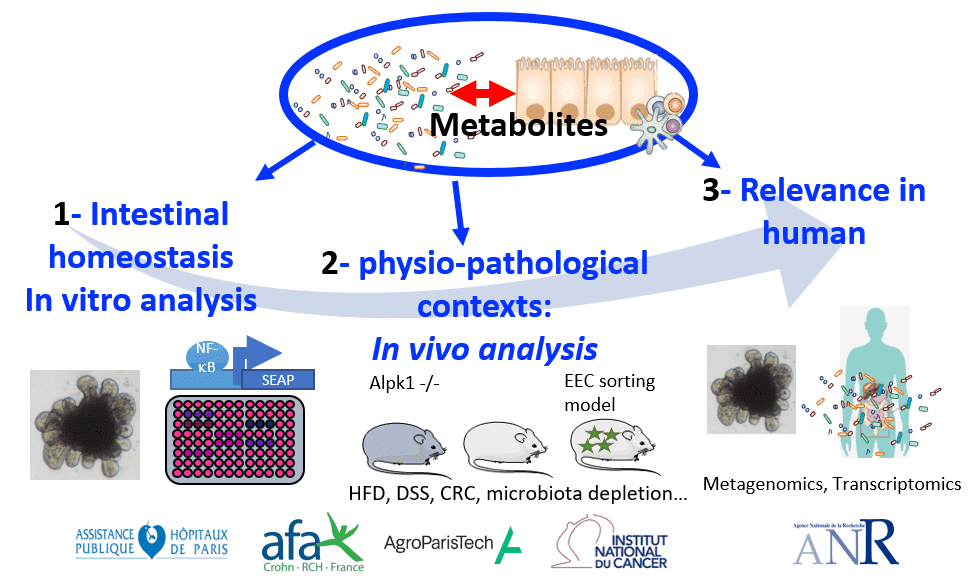
Response of the host to microbial modulations
- Role of innate immune receptors on intestinal homeostasis (obesity, colitis and colorectal cancer).
- Role of the microbiota on enteroendocrine cells plasticity, impact on gut hormones
- Tools used: cell biology (reporter cell lines, CRISPR-Cas9 gene editing, organoids), in vivo analysis (KO and mouse models for cell sorting), transcriptomics, peptidomics
Team members
Alumni
- Liste à puce
- Liste à puce
- Liste à puce
- Liste à puce
- Liste à puce
- Liste à puce
- Liste à puce
Key points
- Martin-Gallausiaux C, Salesse L, Garcia-Weber D, Marinelli L, Beguet-Crespel F, Brochard V, Le Gléau C, Jamet A, Doré J, Blottière HM, Arrieumerlou C, Lapaque N (2024) Fusobacterium nucleatum promotes inflammatory and anti-apoptotic responses in colorectal cancer cells via ADP-heptose release and ALPK1/TIFA axis activation. Gut Microbes 16(1).
- Ngom SI, Maski S, Rached B, Chouati T, Oliveira Correia L, Juste C, Meylheuc T, Henrissat B, El Fahime E, Amar M, Béra-Maillet C (2023) Exploring the hemicellulolytic properties and safety of Bacillus paralicheniformis as stepping stone in the use of new fibrolytic beneficial microbes. Scientific Reports 2023 13:1 13(1):1–16.
- Martin-Gallausiaux C, Garcia-Weber D, Lashermes A, Larraufie P, Marinelli L, Teixeira V, Rolland A, Béguet-Crespel F, Brochard V, Quatremare T, Jamet A, Doré J, Gray-Owen SD, Blottière HM, Arrieumerlou C et al (2022) Akkermansia muciniphila upregulates genes involved in maintaining the intestinal barrier function via ADP-heptose-dependent activation of the ALPK1/TIFA pathway. Gut Microbes 14(1).
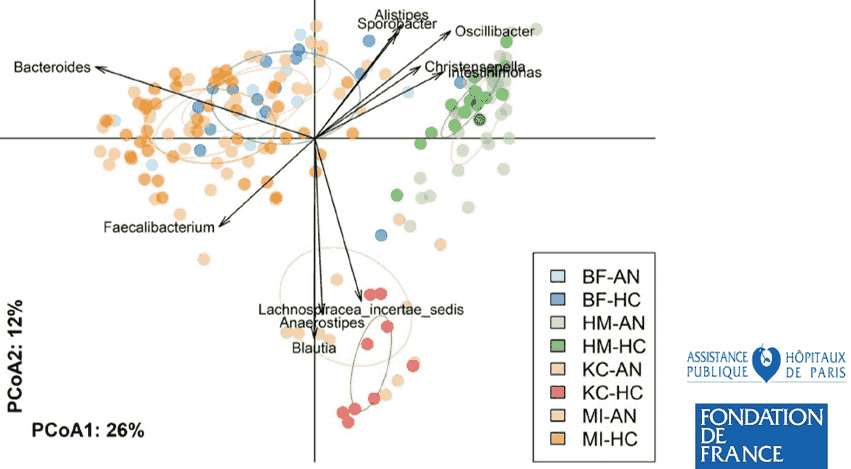
- Mondot S, Lachkar L, Doré J, Blottière HM, Hanachi M (2022) Roseburia, a decreased bacterial taxon in the gut microbiota of patients suffering from anorexia nervosa. European journal of clinical nutrition 76(10):1486–1489.
- Di Lodovico L, Mondot S, Doré J, Mack I, Hanachi M, Gorwood P (2021) Anorexia nervosa and gut microbiota: A systematic review and quantitative synthesis of pooled microbiological data. Progress in Neuro-Psychopharmacology and Biological Psychiatry 106
- van de Guchte M, Mondot S, Doré J (2021) Dynamic Properties of the Intestinal Ecosystem Call for Combination Therapies, Targeting Inflammation and Microbiota, in Ulcerative Colitis. Gastroenterology 161(6):1969-1981.e12.
- Martin-Gallausiaux C, Marinelli L, Blottière HM, Larraufie P, Lapaque N (2021) SCFA: mechanisms and functional importance in the gut. Proceedings of the Nutrition Society 80(1):37–49.
- Van de Guchte M, Burz SD, Cadiou J, Wu J, Mondot S, Blottière HM, Doré J (2020) Alternative stable states in the intestinal ecosystem: proof of concept in a rat model and a perspective of therapeutic implications. Microbiome 8(1):153.
See alll our collection HAL here
External Fundings
- ANR EnteroFructose (2024-2027) : Coordinatrice Véronique Douard
- ANR Gut-Alpk1 (2022-2025) : Nicolas Lapaque
- ANR IMAT (2021-2024) : Coordinateur Maude LeGall (INSERM)
- PLBIO-INCA (2021-2024) : Nicolas Lapaque
- ERC Advanced Homo-Symbosius (2019-2023) : Joël Doré
- EU FP7 Microb-Predict (2019-2026) Coordinateur : Jonel Trebicka (EF CLIF, Barcelone)
- ANR LUMI (2019-2022) Coordinatrice : Sylvia Cohen-Kaminsky (INSERM)
Plusieurs projets collaboratifs avec des industriels sont également en cours DuPont, Sanofi et Roquette.
GRANDS PROJETS passés auxquels nous avons contribué :
- ANR LUMI (2019-2022) Coordinatrice : Sylvia Cohen-Kaminsky (INSERM)
- ANR FunAMetaGen (2015-2019) Coordinateur : Laurent Chene (Entérome)
- ANR PigletBiota (2014-2018) Coodinateur : J Estellé (INRA – Gabi)
- ANR ProteoCardis (2016-2020) Coordinatrice : Catherine Juste
- EU FP7 MetaCardis (2012-2018) Coordinatrice : K Clément (ICAN)
- EU FP7 IHMS (2011-2015) Coordinateur : SD Ehrlich (MGP)
- EU FP7 METAHIT (2008-2012) Coordinateur : SD Ehrlich (MGP)
- EU FP7 Cross-Talk (2008-2012) Coordinateurs : E Maguin (Ife – Micalis) & HM Blottière (FInE – Micalis)
- ANR SUSFLORA (2011-2014) Coordinatrice : C Rogel-Gaillard (INRA – Gabi)
- ANR Surfing (2011-2014) Coordinateurs : G Jan (STLO – INRA) & M van de Guchte (Ife – Micalis)
- ANR SFBIMPRO (2010-2013) Coordinatrice : V Gaboriau-Routhiau (INSERM – Necker)
- ANR EPIFLORE (2013-2016) Coordinatrice : MJ Butel (Université Paris Descartes)
- ANR FunMetaGen (2012-2014) Coordinateur : HM Blottière (FInE – Micalis)
- ANR MicroObes (2008-2012) Coordinateur : J Doré (FInE – Micalis)
- Risk OGM CRYMUC (2012-2016) Coordinatrice : C. Nielsen-Leroux (GME – Micalis)
- INRA MEM MetaScreen (2011-2013) Coordinatrice : G Veronese (LISBP – INRA
- INRA MEM OBOMICS (2012-2014) Coordinatrice : C Juste (FInE – Micalis)